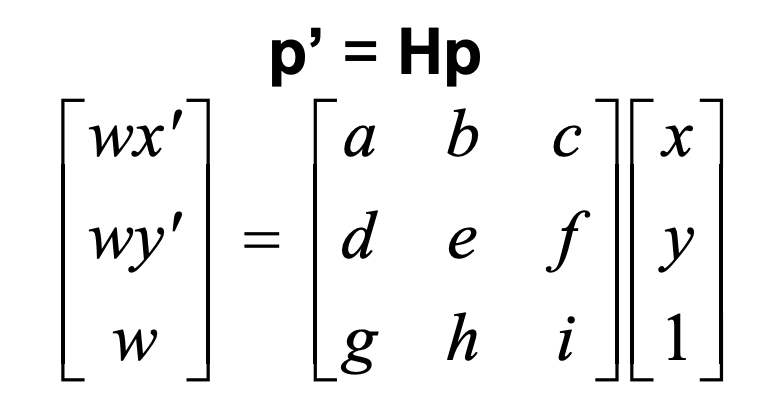Project overview
The first part of the project involved warping images using homographies, and then using homographies to rectify images and combining images into a mosaic. Homographies require 8 degrees of freedom, so we need 4 points (minimum) to solve for a homography matrix.
Homography
We are trying to solve for H, using the formulation below (letting i = 1):
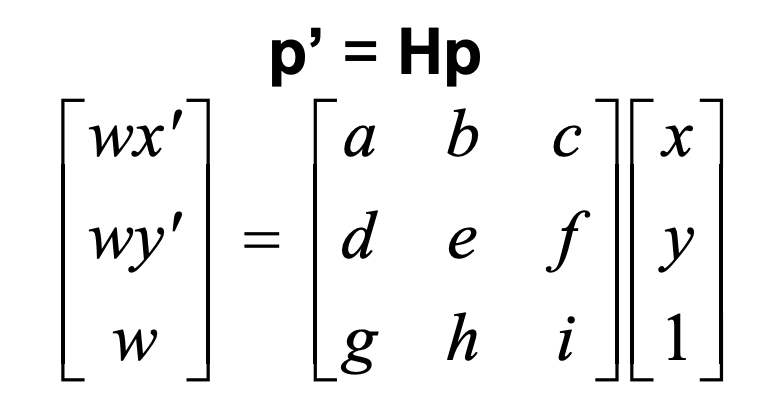
Let's say we are trying to morph from image 1 to image 2. Let (x,y) be some coordinate of image 1, and (x',y') be the corresponding coordinate in image 2. After doing some quick maths, we find that
ax + by + c - gxx' -hyx' = x'
dx + ey + f - gxy' -hyy' = y'
If we have 4 correspondences (minimum), we can solve for all the free variables in H. However, we would like to have more points, and use least squares to fit those points, as humans like myself are pretty bad at clicking corresponding points accurately/consistently.
Warp image result
Here, I warp image 1 to fit the plane of image 2. In order for the warped images to display correctly, they must be translated such that all coordinates are positive.
Image rectification results
These images were rectified by using known square or rectangular coordinates. For the table, I used a 100x100 rectification. For the light switch, I used a 200x300 rectification
Mosaic results
For parts that overlapped, I used a weighted average of both images' pixels, and for the non overlapping parts, I just used the original image pixels.
Conclusions / Coolest things I learned
I thought non-affine transformations would be very difficult, but turns out with one more degree of freedom, we can do morphs! Throughout the entire time we have been learning about image warping, I am starting to get a better understanding on how human vision and perspective works, and how we are able to translate a 2d image on our retina into a 3d perception.
Project overview
Instead of manually selecting correspondence points, we use the MOPS algorithm to automatically detect correspondences, then use RANSAC to obtain a robust homography estimate. The images used here to create the mosaics are the same as in part 1, but in black and white.
Harris Interest Point Detector
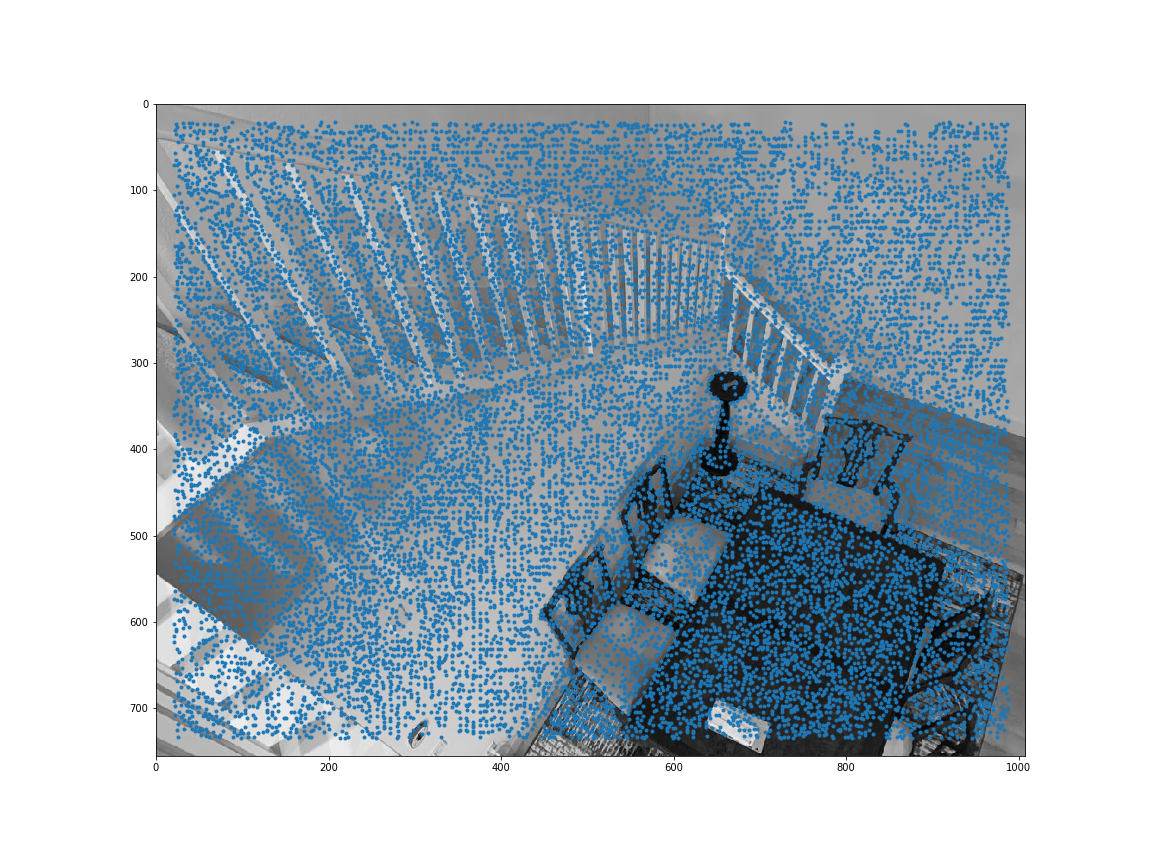
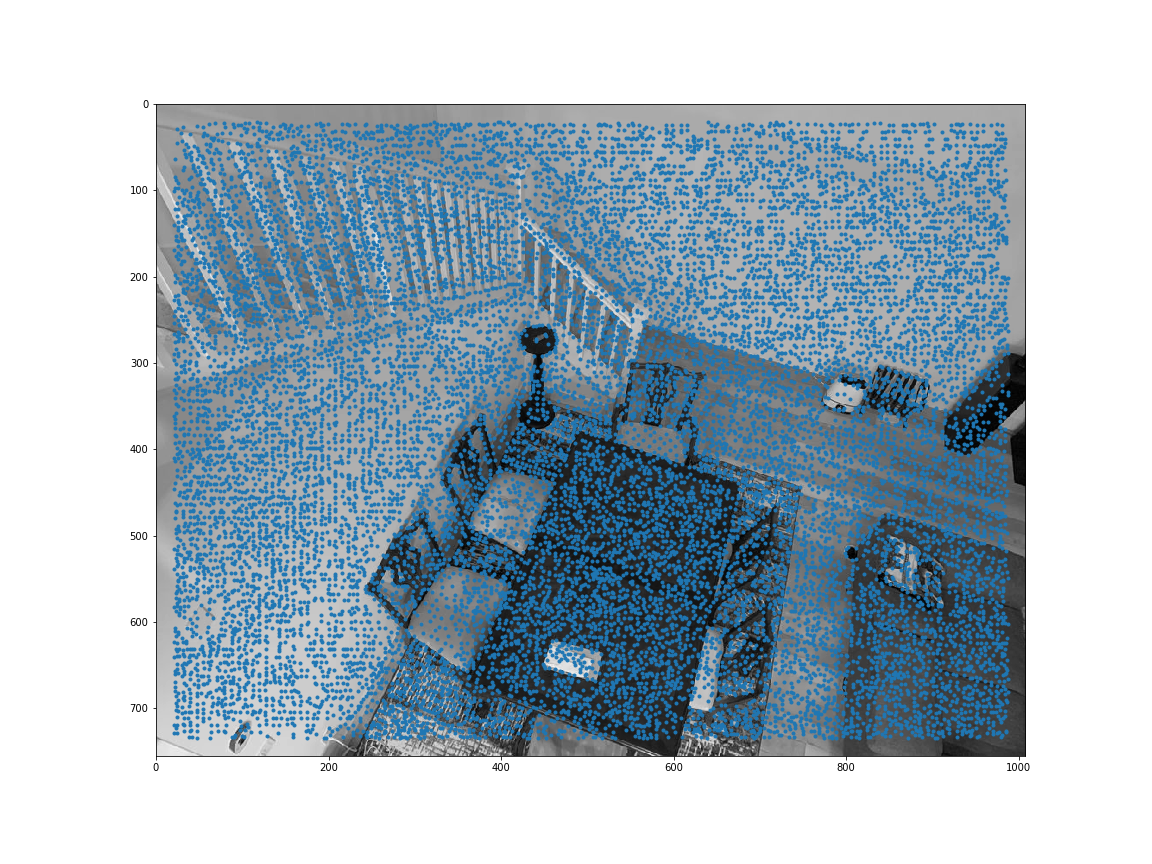
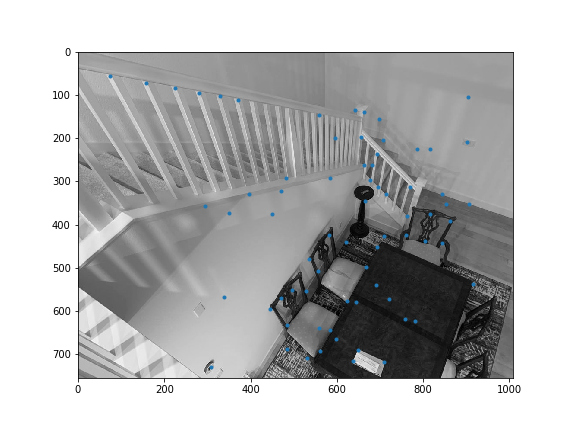
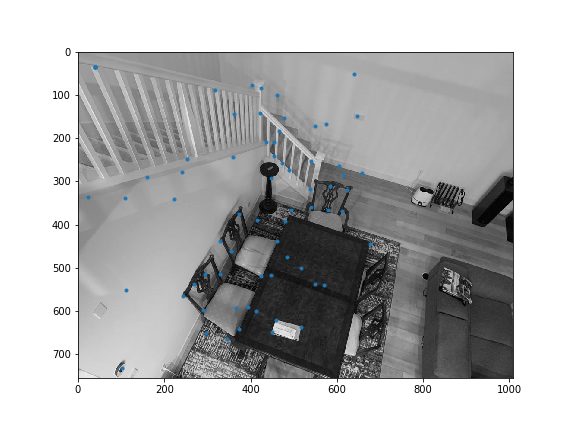
Sample code was provided to obtain all the harris corners. get_harris_corners returns the coordinates of harris corners and the h activation value for all pixels. Here are all the Harris corners overlayed on the initial 2 images I used for mosaicing.
Adaptive Non Maximal Suppression
We want to find the points with the greatest activation evenly distributed on the image. These are more likely to be corners/distinctive points that can be matched among 2 images. My function loops through all harris corners, determines the distance to the closest point that has greater activation, sorts all points by that distance, and then takes the top n=500 points to be our interest points.
Feature descriptor extraction
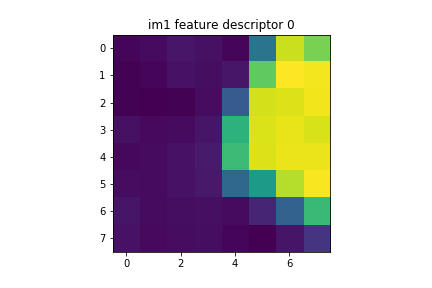
For all interest points, we start with a 40x40 px window around the point to get a feature descriptor. Since we don't want to overfit, we then downsample to an 8x8 px feature descriptor. Finally, we normalize the feature descriptor such that the mean is 0 and the standard deviation is 1.
There are 500 feature descriptors per image (since we had 500 interest points) Here are some examples of feature descriptors.
Feature matching
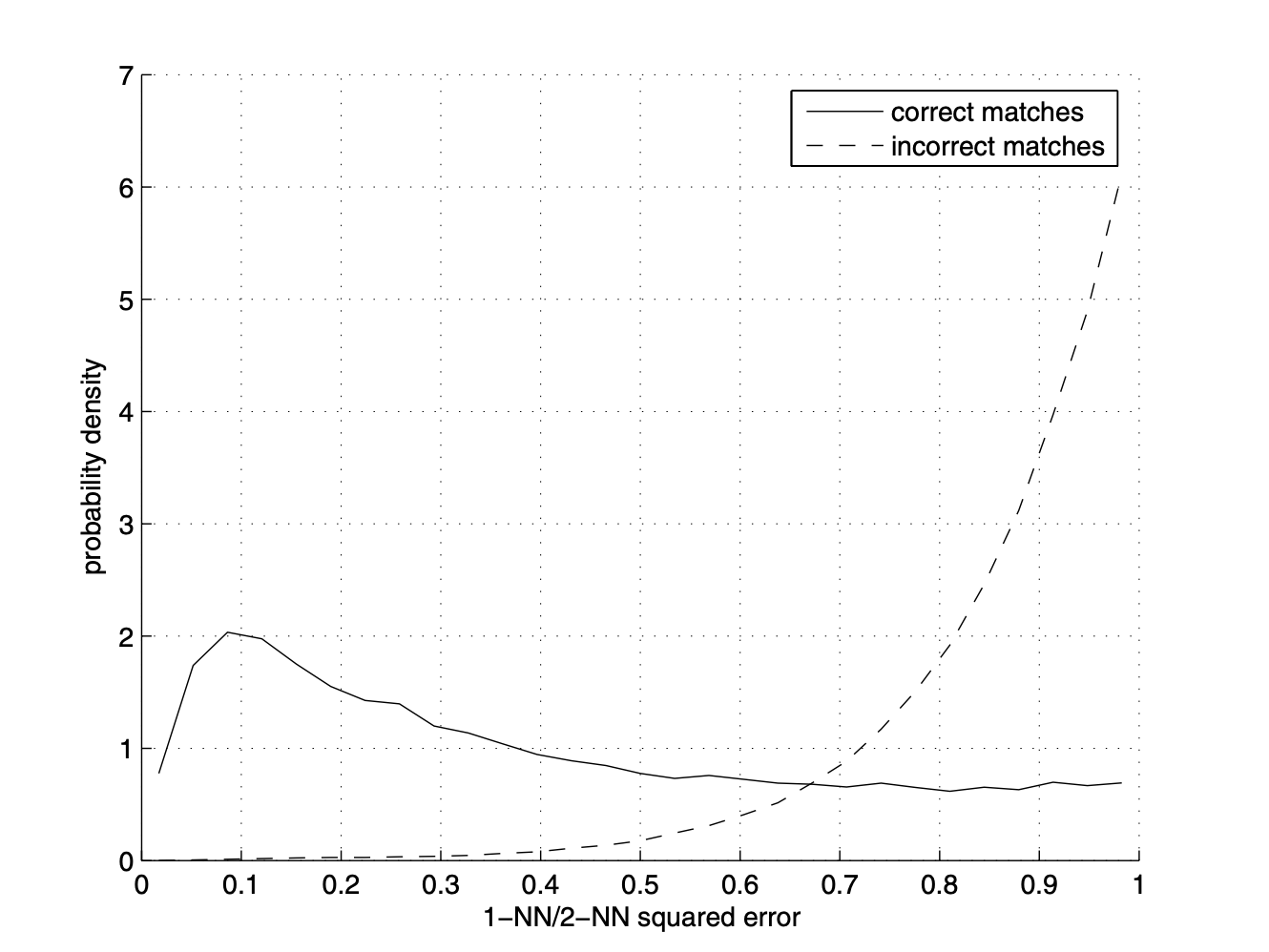
We now try to find the features that match among the 2 images. For each feature for im1, we compute the first nearest neighbor (nn1) and second nearest neighbor (nn2) to a feature for im2. Instead of just comparing the first nearest neighbor, which is prone to a lot of error, we compare each feature's nn1/nn2 ratio. Those with the lower ratios are likelier to be matches, as the difference between the first nearest neighbor distance and second nearest neighbor distance will be large.
All feature pairs that have nn1/nn2 that falls beneath a threshold will be likely matching features. I initially used the chart in the paper to determine the cutoff threshold, then adjusted as needed for my data.
Here are the interest points we kept after doing feature matching. As you might notice, there are still outliers.
RANSAC
In order to address the problem of lingering outliers, we use RANSAC (Random Sample Consensus) to estimate the best fit homography. For a large number of loops (I used 1000), we select 4 random pairs of points and compute the homography matrix. Then, we apply the homography to all point correspondences obtained from feature matching. Next, we compute the number of inliers, which are point pairs such that dist(Hp, p') < epsilon, where epsilon is a small number (ie 0.1). Whichever homography estimate contains the greatest number of inliers will the inliers we use to compute our final homography estimate.The process is also outlined in the slides.
One thing I noticed is sometimes the estimated homography would map every single point in im1 to the same point. This is obviously incorrect, but it does end up with a very large number of inliers, which caused bugs in some rounds. I got around this by ensuring that for the 4 points I selected, if the points H*p are the same, we don't consider that homography matrix H.
Here is the final mosaic for the images
Final mosaics
Comparison between the manually stitched and automatically stitched images (same images as the ones used in part 1). They all did pretty good!
Manually stitched
Automatically stitched
Conclusions / Coolest Thing I Learned
I am surprised by how well the algorithm works, even with so much noise and error in detecting points. The feature detection did not even take into account rotation, and it still managed to find matching features. The coolest thing by far was how the RANSAC algorithm, which is quite simple, was able to determine the best homography despite all the outliers.
Furthermore, I improved my numpy skills which was much needed. I thought I could get away with 2 for loops for adaptive nonmaximal suppression, but that made debugging so difficult because it would take like 10 minutes to run per image. After I got some advice to use np.where magic, it sped up so much and I was very happy. Don't code while tired.